180x Filetype PDF File size 0.13 MB Source: www.alliot.fr
SouthernBlottingand Secondaryarticle
RelatedDNADetection Article Contents
. Introduction
Techniques . ThePrincipleoftheSouthernBlot
. PreparationofDNAPriortoSouthernBlotting
TerenceABrown,UniversityofManchesterInstituteofScienceandTechnology,Manchester,UK . DNABlottingTechniques
. DNA/DNAHybridization
SouthernblottingisatechniquefortransferofDNAmoleculesfromanelectrophoresisgel . TypesofHybridizationProbes
to a nitrocellulose or nylon membrane, and is carried out prior to detection of specific . ApplicationsandLimitationsofSouthernBlotting
molecules by hybridization probing. . Summary
Introduction
Southern blotting is one of the central techniques in ThePrincipleoftheSouthernBlot
molecularbiology.FirstdevisedbyE.M.Southern(1975), TheabilityofnitrocellulosepowderorsheetstobindDNA
Southern blotting results in transfer of DNA molecules, has been known for many years and was utilized in the
usually restriction fragments,from an electrophoresis gel 1950sand1960sinvarioustypesofnucleicacidhybridiza-
to a nitrocellulose or nylon sheet (referred to as a tion studies. In these early techniques the immobilized
‘membrane’),insuchawaythattheDNAbandingpattern DNAwasunfractionated,simplyconsistingoftotalDNA
present in the gel is reproduced on the membrane. During that wasboundtonitrocellulosepowderorspottedontoa
transfer or as a result of subsequent treatment,the DNA nitrocellulose sheet. The introduction in the early 1970s of
becomesimmobilizedonthemembraneandcanbeusedas gel electrophoresis methods that enable restriction frag-
a substrate for hybridization analysis with labelled DNA ments of DNA to be separated on the basis of their size
or RNA probes that specifically target individual restric- promptedthedevelopmentoftechniquesforthetransferof
tion fragments in the blotted DNA. In essence,Southern separated fragments en masse from gel to nitrocellulose
blotting is therefore a method for ‘detection of a specific support. The procedure described by Southern (1975),
restriction fragment against a background of many other involving capillary transfer of DNA from the gel to a
restriction fragments’ (Brown,1999). The restricted DNA nitrocellulose sheet placed on top of it,was simple and
might be a plasmid or bacteriophage clone,Southern effective,and although embellished over the years this
blotting being used to confirm the identity of a cloned original procedure differs only slightly from the routine
fragment or to identify an interesting subfragment from methodstill used in many molecular biology laboratories.
within the cloned DNA,or it might be genomic DNA,in
which case Southern blotting is a prelude to techniques
suchasrestrictionfragmentlengthpolymorphism(RFLP) TheoriginalmethodologyforSouthern
analysis.
InthisarticlethegeneralprinciplesofSouthernblotting blotting
aredescribed,followedbyoverviewsofthemethodologies The original methodology for Southern blotting is
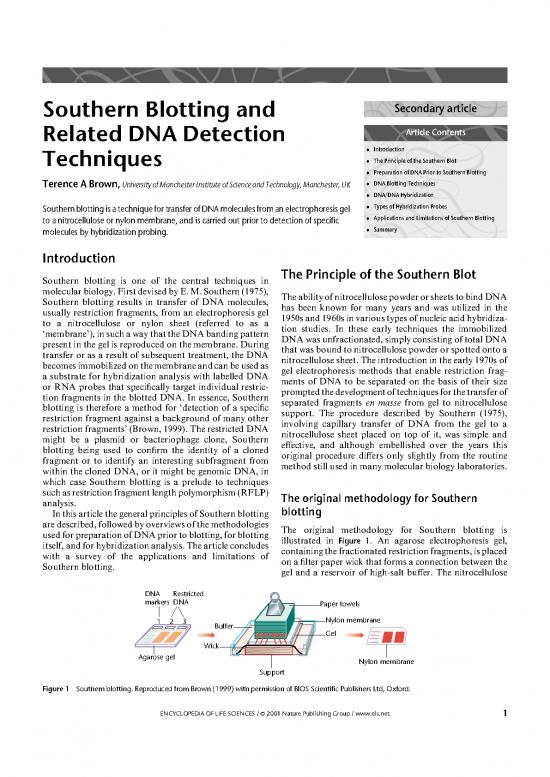
usedforpreparationofDNApriortoblotting,forblotting illustrated in Figure 1. An agarose electrophoresis gel,
itself,andforhybridizationanalysis.Thearticleconcludes containingthefractionatedrestrictionfragments,isplaced
with a survey of the applications and limitations of onafilterpaperwickthatformsaconnectionbetweenthe
Southern blotting. gel and a reservoir of high-salt buffer. The nitrocellulose
DNA Restricted
markers DNA Paper towels
123 Buffer Nylon membrane
Gel
Wick
Agarose gel Nylon membrane
Support
Figure 1 Southernblotting. Reproduced from Brown (1999) with permission of BIOS Scientific Publishers Ltd, Oxford.
ENCYCLOPEDIAOFLIFESCIENCES/&2001NaturePublishingGroup/www.els.net 1
SouthernBlottingandRelatedDNADetectionTechniques
membrane is placed on top of the gel and covered with a capillary blotting system have been proposed but these
towerofpapertowelsthatareheldinplacewithaweight. alternative arrangements are more complicated to set up
Capillary action results in the buffer soaking through the than the simple upward transfer shown in Figure 1,and
filter paper wick,gel and membrane and into the paper none provides any major improvement in speed or
towels. As the buffer passes through the gel the DNA efficiency. Improvements can,however,be achieved by
fragments are carried with it into the membrane,where the use of a noncapillary transfer method,such as
theybecomeboundtothenitrocellulose.Effectivetransfer electroblotting,which employs electrophoresis to move
of fragments up to 15kb in length takes approximately the DNA from the gel to the membrane. This technique
18h,roughlyequivalentto‘overnight’.Theonlytechnical wasoriginally developed for blotting polyacrylamide gels,
complication is the possibility that the buffer bypasses the whose pore sizes are too small for effective capillary
gel by soakingdirectlyfromwicktopapertowels,whichis transferofDNA,andhasalsobeenappliedtoagarosegels
unlikely if the setup is assembled carefully. (Bittner et al.,1980). Electroblotting is more rapid than
capillary transfer but has to be carried out with care
becauseproblemscanarisewithbubbleformationifthegel
MorerecentmodificationstoSouthern getstoohotduringthetransfer.Vacuumblottingisamore
blotting popular choice for agarose gels,a vacuum pressure being
used to draw the buffer through the gel and membrane
ThesystemshowninFigure1isanaccuratedescription of more rapidly than occurs by simple capillary action,
Southernblottingasstillcarriedoutinmanylaboratories, enabling the transfer time to be reduced to as little as
but various modifications have been introduced over the 30min.Attemptshavealsobeenmadetodispenseentirely
yearstoimprovetheefficiencyofDNAtransferfromgelto with Southern blotting and carry out hybridization
membrane. The major improvement has been the intro- analysis directly with the electrophoresis gel,immobiliza-
ductionofnylonmembranes,whichhavethreeadvantages tion of the DNA being achieved by drying the gel so that
over their nitrocellulose counterparts. First,nylon mem- theDNAbecomestrappedinthedehydratedgelmatrixor
branes are less fragile than nitrocellulose sheets,the latter boundtoasolidsupportontowhichthegelisplacedprior
tending to crack if handled roughly during Southern to drying. These techniques have proved useful for some
blotting,andusuallydisintegratingifattemptsaremadeto applicationsbutsufferfromhighbackgroundsignalsafter
carry out more than two or three hybridization analyses hybridization analysis.
withthesameblot.Nylonmembranescannotbedamaged
byhandlingandasingleblotcanberehybridizeduptoten
times,this limit being due not to eventual breakage of the PreparationofDNAPriortoSouthern
membrane but to the gradual loss of the blotted DNA
during repeated hybridizations. The second advantage of Blotting
nylon membranes is that under certain conditions (a
positively charged membrane and an alkaline transfer PreparationofgenomicDNA
buffer)thetransferredDNAbecomescovalentlyboundto
the membrane during the transfer process. This is not the ThetechniquesusedtoprepareDNAforSouthernblotting
case with a nitrocellulose membrane,which initially binds depend on the type of DNA that is being studied. For
DNAinasemipermanentmanner,immobilizationoccur- genomic DNA,the objective is to obtain molecules that
ring only when the membrane is baked at 808C. Transfer have not become extensively fragmented by random
onto a positively charged nylon membrane can therefore shearing during the extraction process,so that specific
reduce the possible loss of DNA that might occur by restriction fragments of 20kb and more can be obtained.
leaching through the membrane during the blotting Cellsmustthereforebebrokenopenunderrelativelygentle
process; it is also quicker,the transfer time being reduced conditions. For tissue culture cells and blood samples
from18hto2h.Finally,nylonmembranesefficientlybind incubation in a buffer containing a detergent such as
DNA fragments down to 50bp in length,whereas sodium dodecyl sulfate (SDS) is usually sufficient to
nitrocellulosemembranesareeffectiveonlywithmolecules disrupt the cell membranes and release high-molecular
longer than 500bp. Nitrocellulose has not,however,been weightDNA.Bacteria,whicharesurroundedbyacellwall,
completely superseded because it has one significant can be gently broken open by treatment with a combina-
advantage compared with nylon membranes: a reduced tion of lysozyme and ethylenediaminetetraacetate
amount of background hybridization,especially with (EDTA). Lysozyme is an enzyme that degrades some of
probes that have been labelled with nonradioactive thepolymericcompoundspresentinthebacterialcellwall,
markers. andEDTAchelatesmagnesiumionsthatarerequiredfor
Other changes to the original Southern blotting proce- integrity of the polymeric structure. Subsequent addition
dure have been introduced to speed up and improve the of a detergent causes the cells to burst. Other microorgan-
efficiencyofthetransfer.Changestothearchitectureofthe ismswithtoughcellwalls,suchasyeastandfungi,canalso
2 ENCYCLOPEDIAOFLIFESCIENCES/&2001NaturePublishingGroup/www.els.net
SouthernBlottingandRelatedDNADetectionTechniques
be treated with appropriate cell wall-degrading enzymes, uptheSouthernblot.Thepretreatmenthastwoobjectives.
and the same is true for plant cells though the latter are First,it is desirable to break the DNA molecules in
usually broken open by physical grinding of tissues that individual bands within the gel into smaller fragments,
have been frozen in liquid nitrogen. This causes some because smaller fragments transfer more quickly than
fragmentation of the DNA but provides a much higher larger ones. This is achieved by soaking the gel in
yield. 0.25molL21 HCl for 30min,which results in a small
After cell disruption,genomic DNA extraction proce- amount of depurination – cleavage of the b-N-glycosidic
dures continue with steps aimed at removing the major bond between purine bases (adenine or guanine) and the
biochemicalsotherthanDNApresentintheinitialextract. sugar component of their nucleotides – which is followed
Aprotease such as proteinase K might be included in the by decomposition of the sugar structure and breakage of
buffer used for cell disruption,in order to begin the the polynucleotide chain. The second pretreatment is with
degradationofproteinsintheextract,butdeproteinization an alkaline solution that denatures the double-stranded
is routinely carried out by phenol extraction,the addition DNAmoleculesbybreakageoftheir hydrogen bonds,so
of phenol or a 1:1 mixture of phenol and chloroform the molecules become single-stranded. This aids their
resulting in precipitation of proteins. After centrifugation, transferandsubsequentbindingtothemembrane,andalso
the precipitated proteins migrate to the interface between ensures that after binding the base-pairing components of
the organic and aqueous phases,whereas the nucleic acids thepolynucleotidesareavailableforhybridizationwiththe
remain in the aqueous phase. For plant extracts,which probe.
containgreaterquantitiesofcarbohydratesthanarefound Ifanitrocellulosemembraneisbeingusedthenthealkali
in most animal and microbial cells,an additional purifica- pretreatment is followed by neutralization of the gel by
tion involving the detergent cetyltrimethylammonium soaking in a Tris-salt buffer,this step being essential
bromide (CTAB) is frequently used. This compound because DNA does not bind to nitrocellulose at a pH of
specifically binds nucleic acids,improving their recovery greater than 9.0. The Southern blot is then set up,as
duringphenolextraction.Mostextractionproceduresalso illustrated in Figure 1,with a high-salt transfer buffer,
include digestion of RNA with ribonuclease and a final usually the formulation called ‘20 SSC’,which
treatment with ethanol,which precipitates the remaining comprises 3.0molL21 NaCl (salt) and 0.3molL21
nucleic acid polymers,enabling ribonucleotides and other sodium citrate. The same buffer can be used for transfer
low-molecular weight contaminants to be removed. The to a nylon membrane,butwithapositively chargednylon
precipitatedDNAisdriedandthenredissolvedinwateror membraneanalkalinetransferbuffer(0.4molL21NaOH)
a Tris–EDTAbuffer. is used because,as described earlier,this results in
immediate covalent attachment of the transferred DNA
PreparationofplasmidorbacteriophageDNA to the membrane. With this type of transfer the alkali
pretreatmentisunnecessary.Theblotisthenleftforatleast
Specialtechniquesmustbeusediftheobjectiveistoobtain 18hforahigh-salt transfer,or 2h for an alkaline blot.
plasmid or bacteriophage DNA from bacterial clones. After blotting,the transfer setup is dismantled and the
Plasmid DNA can be obtained by any one of several membranerinsedin2 SSCandlefttodry.Iftheblothas
techniques that exploit the physical differences between been made onto a nitrocellulose or uncharged nylon
plasmids and bacterial genomic DNA,plasmids being membrane,then the DNA is only loosely bound to the
small supercoiled molecules whereas genomic DNA,after membrane at this stage. More permanent immobilization
cell disruption,is present as long,linear fragments. A mustthereforebecarriedout,eitherbybakingat808Cfor
popularmethodinvolvestreatmentofthecellextractwith 2h,which results in noncovalent but semipermanent
alkali,which precipitates the linear DNA but leaves attachment of DNA to a nitrocellulose membrane,or
supercoiled plasmids in solution. Bacteriophage DNA, UVirradiation,which results in covalent attachment of
for example recombinant l or M13 vectors,is usually DNAtoanylonmembrane.
obtaineddirectlyfrombacteriophageparticlessecretedby
infected bacteria,the purification process being simplified
by the fact that these particles consist solely of DNA and DNA/DNAHybridization
protein.
Hybridization analysis is based on the principle that two
polynucleotideswillformastablehybridbybase-pairingif
DNABlottingTechniques their nucleotide sequences are wholly or partly comple-
mentary.AspecificrestrictionfragmentinaSouthernblot
AfterthepurifiedDNAhasbeentreatedwithoneormore canthereforebedetectedifthemembraneisprobedwitha
restriction endonucleases it is fractionated by agarose gel second,labelled DNA molecule that has the same,or
electrophoresis and the gel then pretreated prior to setting similar,sequence as the fragment being sought. We will
ENCYCLOPEDIAOFLIFESCIENCES/&2001NaturePublishingGroup/www.els.net 3
SouthernBlottingandRelatedDNADetectionTechniques
examinethetypesofhybridizationprobethatcanbeused a 10- to 100-fold increase in sensitivity can be achieved
later; first we will survey the methodology used for (Amasino,1986).
hybridization analysis. Thesecondcriticalfactorthatmustbeconsideredduring
thehybridizationstepisthespecificityofthereaction.Ifthe
probeDNAhasbeencarefullychosenthenitwillcontaina
PrehybridizationofaSouthernblot regionthatiscompletelycomplementarytoallorapartof
theblottedrestrictionfragmentthatisbeingsought.Ifthis
Hybridization analysis is carried out by soaking the hybridizing region in the probe is not completely com-
Southern blot in a buffer containing the hybridization plementarytothetarget,thenitwillatleasthavearegionof
probe,usually in a tube that is constantly rotated so all strong similarity so that a stable hybrid can form. The
parts of the membrane are exposed to the probe,or problemisthattheprobealsohasthepotentialtohybridize
alternatively in a sealed plastic bag that is placed on a to any other blotted DNA fragments with which it has
shaker.Hybridizationiscarriedoutintwostages.First,the partial complementarity. The hybridization experiment
membraneisprehybridizedinasolutiondesignedtoblock will never give an unambiguous signal if the probe is more
theunusedDNAbindingsitesonthemembranesurface.If similartoasecondrestrictionfragmentthanitistotheone
this step is omitted then the probewillbindnonspecifically beingsought,butproblemswithnonspecifichybridization
to the surface of the membrane and the signal resulting can still arise even if the best match is with the specific
fromhybridizationtothespecificrestrictionfragmentwill target. This means that the hybridization step must be
bedifficultifnotimpossibletoidentify.Theprehybridiza- carried out at a ‘stringency’ that results in the specific
tion solution therefore contains nonbiological polymeric probe–target hybrid remaining stable while all other
compounds such as polyvinylpyrrolidone and/or biologi- hybrids are unstable,the stringency being determined by
cal polymers such as Ficoll (a carbohydrate-based the composition of the hybridization buffer and the
compound),bovine serum albumin or dried milk. DNA temperatureatwhichtheexperimentiscarriedout.Buffer
from an organism unrelated to the one whose DNA has composition is relevant because hybrid stability is depen-
been blotted can also be used (salmon sperm DNA is a dentontheionicstrengthandthepresenceofdestabilizing
popular choice). Prehybridization takes between 15min agents,suchasformamide,whichdisrupthydrogenbonds.
and3hat688C,dependingonthetypeofmembrane. Temperature is relevant because the melting temperature
(T ,thehighesttemperatureatwhichthehybridisstable)
m
of a fully base-paired hybrid is higher than that for one in
Thehybridizationandwashingsteps whichsomebasepairshavenotformedbecausetheprobe
andtargetDNAsarenotfullycomplementary.Formation
The second stage is the actual hybridization,which is of the desired hybrid,and destabilization of nonspecific
carried out in a high-salt buffer containing a detergent, hybrids,can therefore be achieved by utilizing an appro-
usually2 SSC11%SDS.Twoissuesarecriticalatthis priate combination of buffer composition and hybridiza-
stage of the experiment. First,enough probe DNA must tion temperature. A number of different strategies are
hybridize to the target restriction fragment to produce a possible. If the probe is 4100bp in length,for example a
clear signal that can be discerned by the detection system clonedrestrictionfragment,thentheinitialhybridizationis
appropriateforthelabelcarriedbytheprobe.Forthemost usually carried out at 688C in a high-salt buffer,this
demanding applicationsuch as detection of a single copy representing highly stringent conditions under which only
gene in human genomic DNA,achieving sufficient stable hybrids are expected to form,with very little if any
sensitivity might be a problem even if the maximum nonspecific hybridization. With this strategy,washing
amountofgenomicDNAisloadedontotheelectrophor- steps that are carried out after hybridization are designed
esis gel (in practice,about 10mg of DNA per lane) and the simply to remove the nonhybridized probe. However,if a
probe has been labelled to its maximal specific activity short oligonucleotide probe is used (15–25 nucleotides in
9 21 32 length),then the hybridization step is usually carried out
(410 dpmmg fora Pradioactivelabel).Increasingly
greater problems will be encountered if a radioactive label under conditions of low stringency (typically at a
32 35 temperature several degrees below the calculated T for
other than P is used,such as S,which is generally m
suitable only for probing less complex DNAs such as thedesiredhybrid),sothatallpotentialhybrids,including
restricted plasmid or bacteriophage clones,or if a nonspecific ones,are able to form. Specificity is then
nonradioactive probe is employed. Under these circum- achievedbyaseriesofwashesatincreasingtemperaturesso
stances some increase in sensitivity can be obtained by that,hopefully,only the desired hybrid remains at the end
including an inert polymer such as 10% dextran sulfate or of the procedure.
8%polyethyleneglycol6000inthehybridizationsolution. After washing,the membrane is subjected to the
Thesepolymersarethoughttoinducetheprobemolecules detection procedure appropriate for the label that has
to form networks so that greater amounts of DNA been used,for example autoradiography for a radioactive
hybridize to the target sites on the membrane. In practice, label. Subsequent reprobing is possible if the membrane is
4 ENCYCLOPEDIAOFLIFESCIENCES/&2001NaturePublishingGroup/www.els.net
no reviews yet
Please Login to review.