211x Filetype PPTX File size 0.77 MB Source: s3-eu-west-1.amazonaws.com
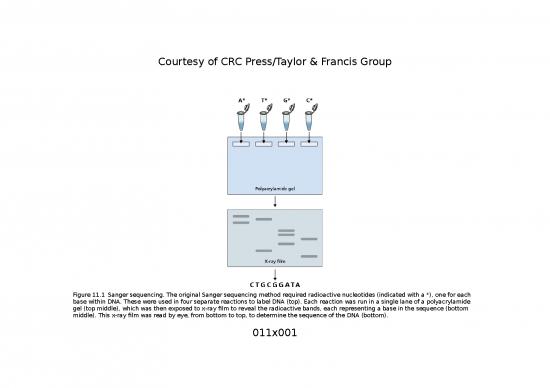
Courtesy of CRC Press/Taylor & Francis Group
Figure 11.2 Sanger sequencing with fluorescent nucleotides. Rather than using four separate radioactive nucleotides in the sequencing
reactions, use of nucleotides each labeled with a different type of fluorescence reduced both the number of reactions (top) and the number
of lanes on the polyacrylamide gel (middle). Fluorescence also enabled automated detection (bottom), reducing human error in reading
bands on an x-ray film as previously (see Figure 11.1).
011x002
Courtesy of CRC Press/Taylor & Francis Group
Figure 11.3 Shotgun sequencing. This methodology, applied to sequencing whole-bacterial genomes, involves first randomly dividing the
genome into fragments that are cloned into a library. The order to the fragments as they were in the genome is lost. As the library is
sequenced, it produces overlapping sequence data that is joined computationally to generate contiguous sequence data.
011x003
Courtesy of CRC Press/Taylor & Francis Group
Figure 11.4 Regions of difference between bacterial genomes. Comparative genomics, evaluating differences and similarities between two
genome sequences (top and bottom), shows that there are some regions in one genome that are not in the other (blue), and vice versa
(red).
011x004
Courtesy of CRC Press/Taylor & Francis Group
Figure 11.5 Next-generation sequencing principles. Most next-generation sequencing technologies use sequencing by synthesis, as established in the
Sanger sequencing method, but with modifications. DNA is fragmented and sequencing technology specific adapter sequences are ligated to the ends of the
fragments. The adapters are used to attach the fragments to a solid surface (bead or flowcell), as primers for amplification of the fragments, and to initiate
sequencing of the fragments. As nucleotide bases are incorporated into the DNA, the identity of each incorporated base is determined via fluorescence or
changes in pH within the reaction vessel.
011x005
Courtesy of CRC Press/Taylor & Francis Group
Figure 11.6 Repetitive regions complicate assembly. If a genome contains, for example, six repetitive regions that are identical to one
another (A, B, C, D, E, and F), this can confound attempts to assemble sequencing fragments. The fragment shown could belong in any of
the six regions of the genome.
011x006
no reviews yet
Please Login to review.